Submitted by GAtherton on 9 November 2016
Back in genomic pre-history (prior to 2000) DNA was sequenced by attaching coloured markers on the end of the DNA fragment that you wanted to read – each base had a different colour so you could separate the resulting DNA reads by length and then read base by base according to colour – this is known as chain termination sequencing. This could be done by machine and computer and thus became relatively cheap compared to its predecessor manual methods and very accurate way to read the sequence of a piece of DNA. This technique was routinely used for many years to sequence a few hundred base pairs up to a few thousand.
However this technique has a drawback – it is still quite slow so if you want to sequence an entire genome (i.e. made up of millions of base pairs) and thus the cost of genome sequencing was high.
Advances in sequencing technology have forced these costs down and sequencing has sped up steadily during 1990’s and 2000’s. High throughput sequencing techniques have now progressed from chain termination sequencing (cost estimated at $2400 per million base pair reads, up to 900bp per run, each run taking up to 3 hours) to Nanopore sequencing (cost estimated at $0.11 per million base pair reads, >5400bp per run, 1 min per run). Crucially Nanopore sequencing can read 4.4 million sequences, each of 5400bp per run!! This means that entire large genomes can be sequenced in a single run. Error rates are higher than earlier methods but repeat runs improve this and the technology will improve.
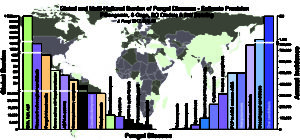
The first fungal genome to be sequenced using Nanopore technology has just been announced demonstrating that this technology can be used for fungi, opening the way to larger scale genome sequencing of many more fungal genomes – for example we will be able to sequence collections of individual clinical isolates of Aspergillus fumigatus in hours/days rather than months giving clinicians virtually real time complete genome information about the infecting fungus that they are treating. Such information could be used quickly to tailor treatment if the patient has an acute infection, or might be used longitudinally to track any changes in DNA sequence as a fungus adapts to its host during chronic infection. In short this has the potential to be both a clinical and research tool which will benefit the diagnosis and treatment fo fungal infections immensely.
News archives
-
Title
Date